# The Variant Page
The variant page provides in-depth information on the variant and can be accessed by clicking on the variant information links in the variant detail panel or HGVSc / HGVSp columns. Alternatively, you can browse this page for any variant by clicking on the “Variants” in the left pop-up menu and searching for the variant of your interest. You can reach the dedicated page for the variant of your interest by clicking on the “Details” button.
# The ACMG Pathogenicity Calculator
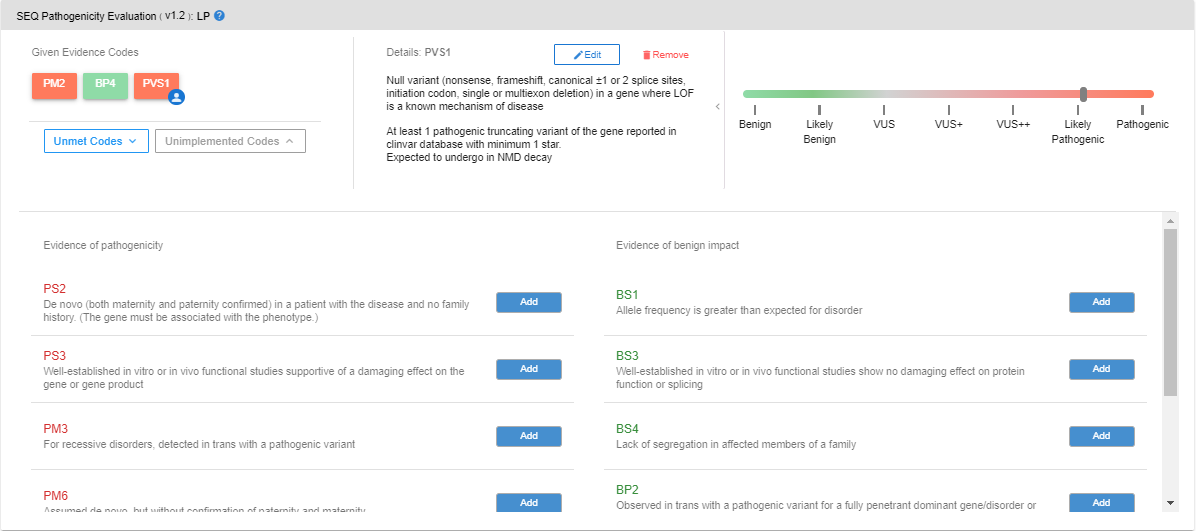
In this section, you can see the ACMG evidence codes assigned to the variant, read their explanations, add/remove evidence codes, and see the changes in the pathogenicity prediction.
In the “Given Evidence Codes” section, you can see which codes are assigned to the selected variant. You can see the explanation in the “Details” section by clicking on these codes. You can see the interactive pathogenicity scale on the right-hand side. If you wish to remove any particular evidence code, you can click on the "Remove" button. The pathogenicity scale will be updated according to the remaining codes.
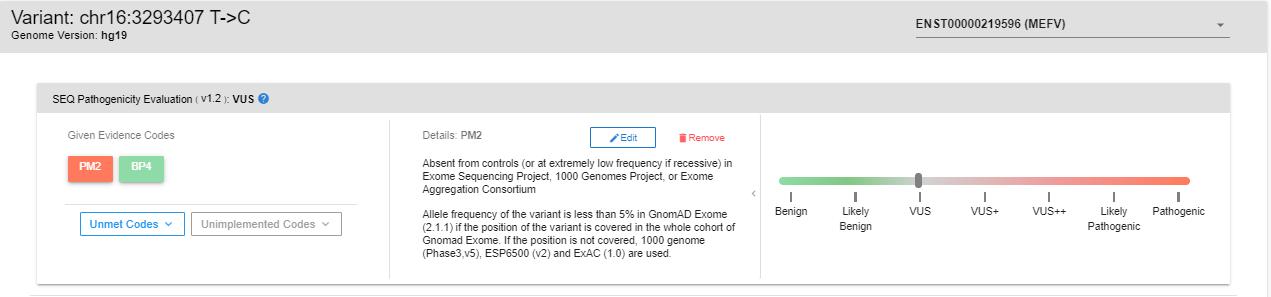
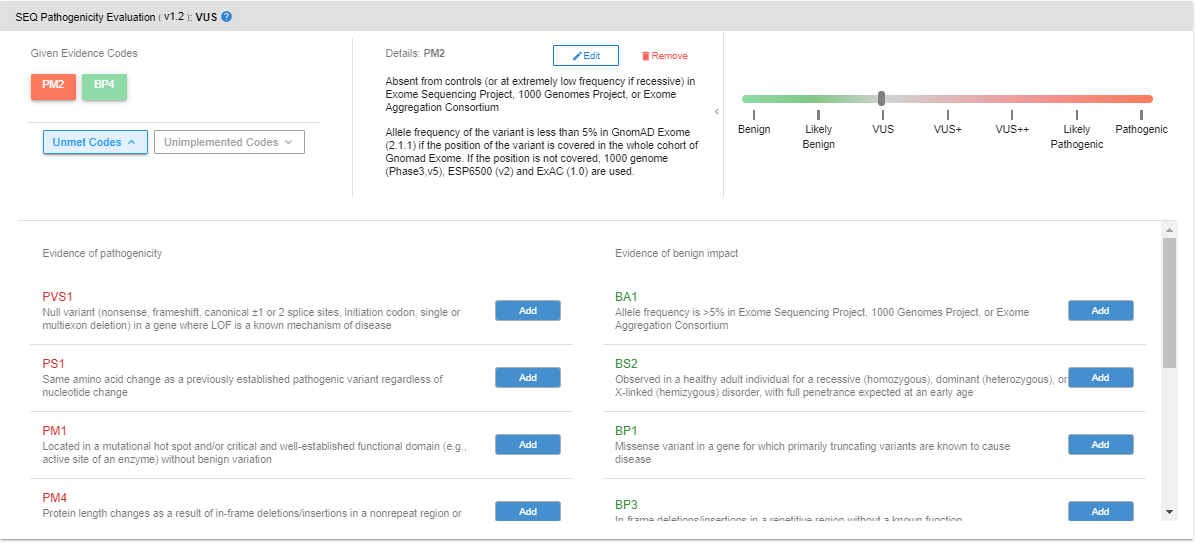
By clicking on the “Unmet Codes” button, you can see the list of evidence codes that are bioinformatically analyzed but not assigned to the variant. You can add such unassigned codes by clicking the “Add” button.
When you add an evidence code, you will be asked to choose the pathogenicity level of the code. The default value declared in the ACMG guideline will be automatically assigned. You can change the strength of the code, should you choose to. You can see the changes in the pathogenicity scale in this pop-up screen.
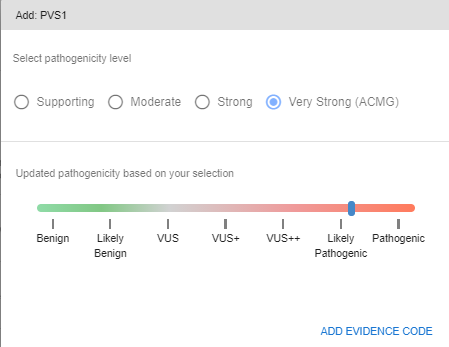
When you add a previously unassigned evidence code, it will be marked for easier recognition.
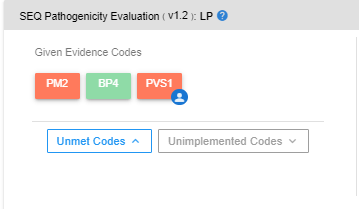
You can also list the codes that cannot be bioinformatically analyzed with the provided data by clicking the “Unimplemented Codes” button. Following the same procedure, you can add unimplemented codes to the variant of your interest.
# Surrounding Sequence
This section shows the base change(s), ten bases up and downstream of the variant, and the genomic coordinates.

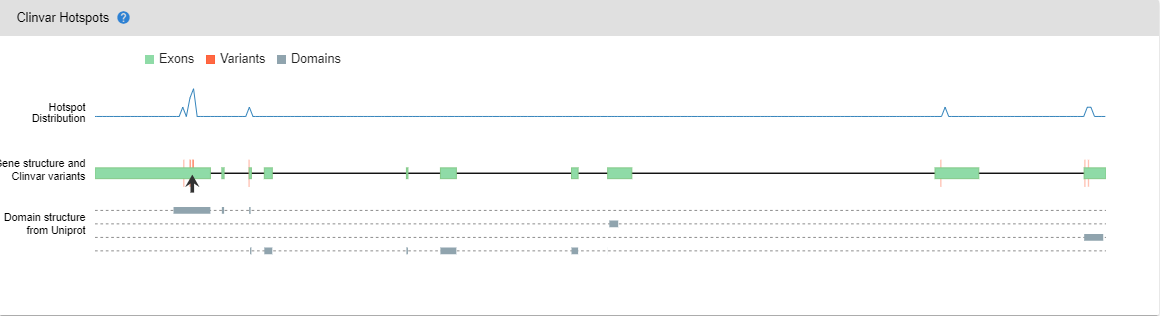
# Clinvar Hotspots
This graph shows the distribution of pathogenic and likely pathogenic entries in the ClinVar database with the gene and domain structures.
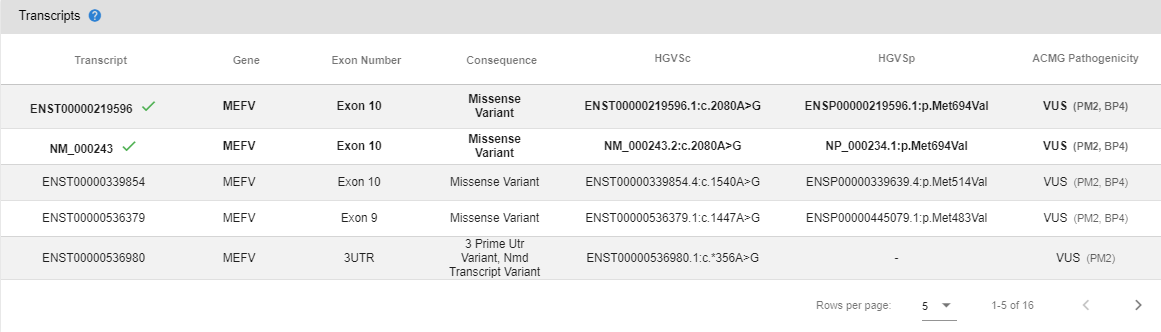
# Transcripts
In this section, you can see the reference and alternative transcripts affected by the variant, their genes, exon numbers, consequences, HGVSc and HGVSp annotations, and ACMG pathogenicity predictions.
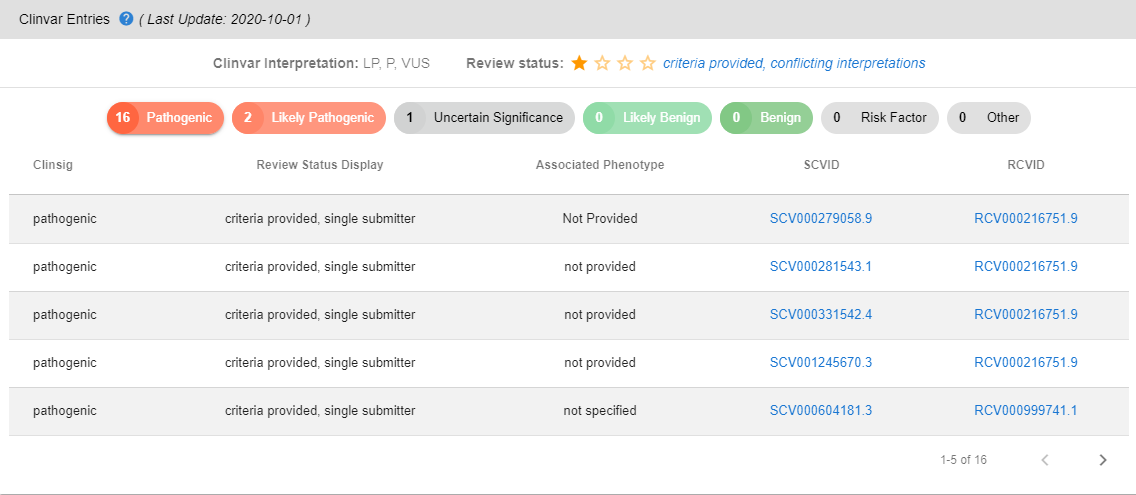
# Clinvar Entries
Here, you can find the ClinVar interpretation, review status, and number of entries in each pathogenicity tier. You can click on the pathogenicity identifiers to create a list with the corresponding entries. You can click on SCVID and RCVID identifiers to be redirected to corresponding pages in the ClinVar database.
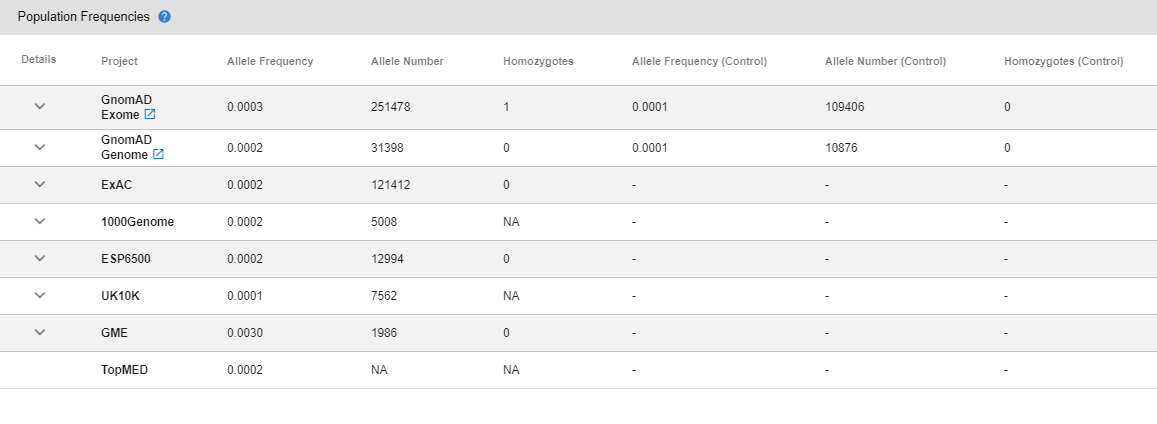
# Population Frequencies
Frequency information from major genome projects is listed in this section. You can see the population stratifications by clicking on the arrow icon for each project if such information is available.
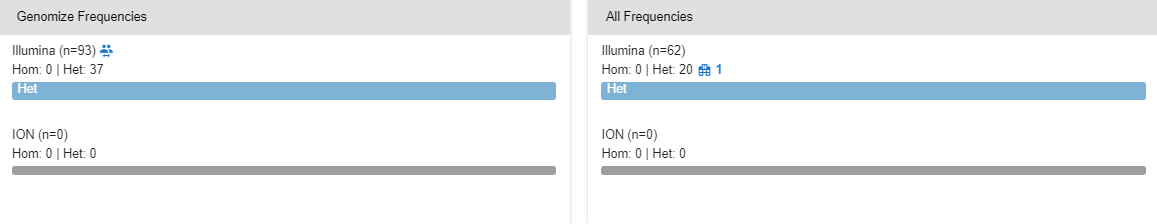
# Site and “All” Frequencies
In these sections, you can see the frequency of the variant within the samples uploaded by your site and within all the samples uploaded to the SEQ Platform by any of its users. In the “Site Frequencies” section, you can click on the icon to list the samples in which this variant is detected. You can also click on these samples to reach the respective variant page of these samples.
In the “All Frequencies” section, you can see the number of sites that have already observed this variant. You can click on the icon to request contact with these sites.
# Literature Information
This section lists all relevant publications with direct links to the PubMed page of the publication. This list is created in real-time.
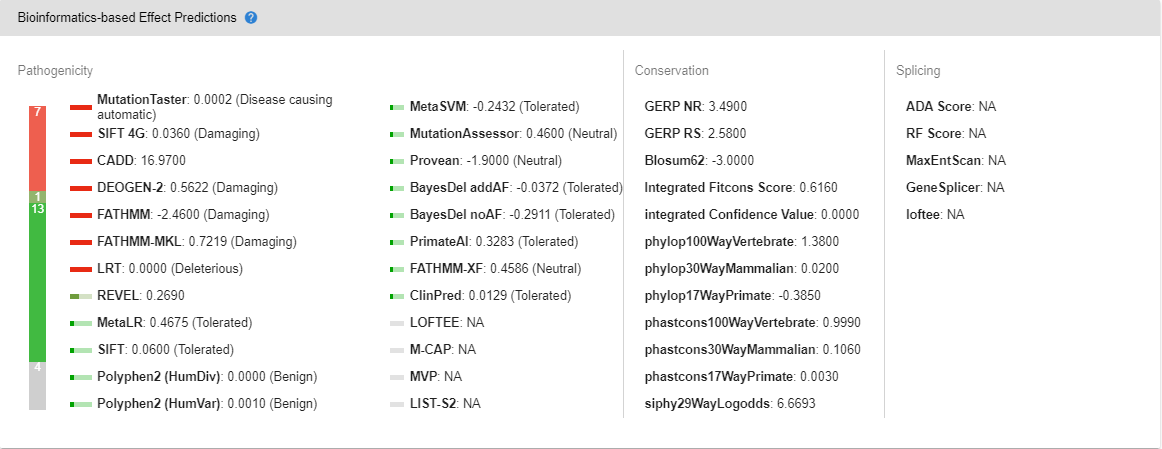
# Bioinformatics-based Effect Predictions
You can find the bioinformatics-based pathogenicity, conservation and splicing predictions from various tools with visually distinct representations to aid in your analyses.