# SEQ genotype/phenotype/disease database
The SEQ platform enables you to build a phenotype/genotype/disease database while you are performing analysis. SEQ calculates the number of samples in which the position is covered as well as the occurrence of the variant. Therefore, it enables the calculation of accurate frequency, which could be used for eliminating repetitive variants due to a) regional polymorphism, b) kit specific false positives, or c) bioinformatics algorithm-specific repetitive variants.
Please note that the generation of the real-time phenotype/genotype/disease database is only available for the fastq files and
not for the vcf files.
The allele frequencies can be visualized in different parts of the platform as follows:
- Allele frequency, can be visualized in the one of the columns in the SEQ sample analysis page as marked below.
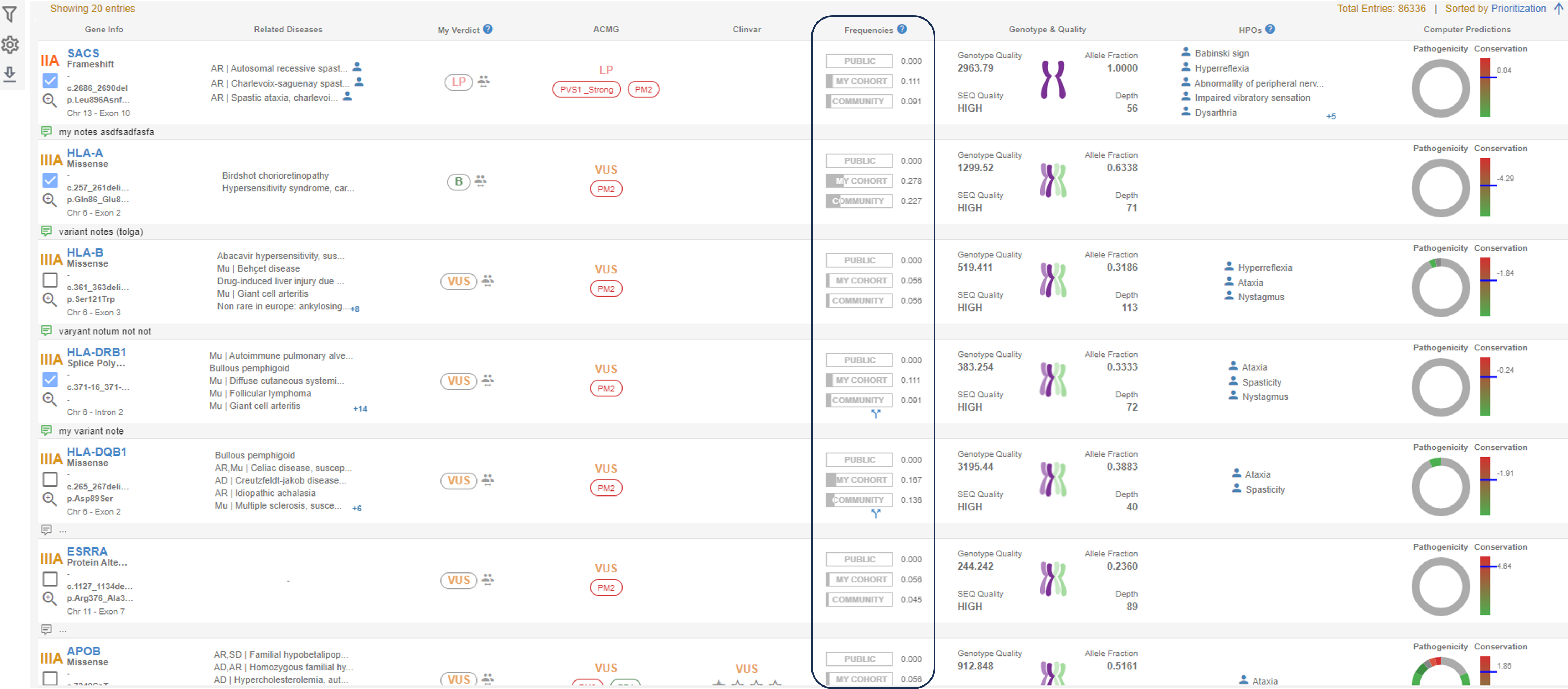
- Genotype frequency segregated by machine type can be visualized upon clicking on the frequency information in any variant. The "Frequencies" panel will open at the bottom of the page.
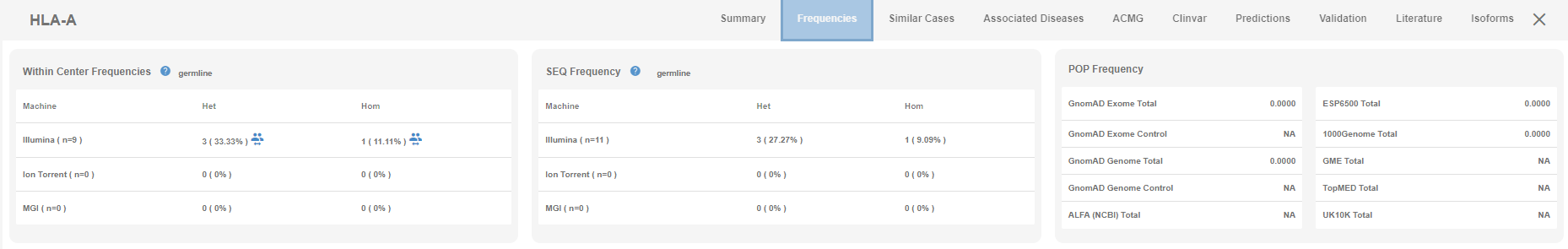
Genotype frequency segregated by the machine and the kit used can be visualized under the variant page along with the more detailed public cohort frequencies.
You can find phenotype and disease associations with the variant in your sample cohort in their respective sections in the information field at the bottom of the page.