# Quality Control
# Amplicon coverage
The Amplicon coverage provides minimum and maximum coverages, and coverage percent (with 1X depth threshold) for the kit’s target regions.
These values can be sorted. In addition, filtering by gene symbol and coverage percent is available.
# Gene coverage
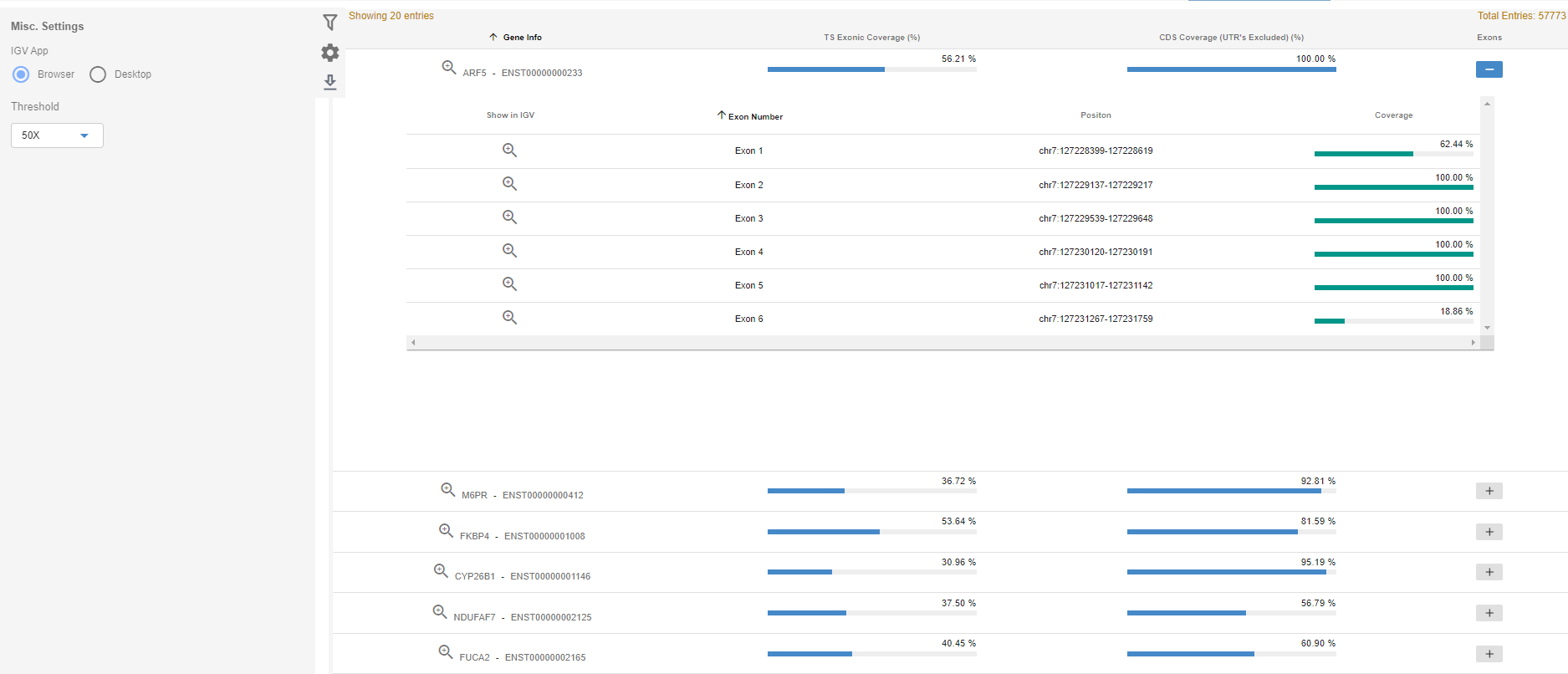
The Gene coverage section provides coverage values for reference transcripts of the genes provided in the selected gene annotation source (Ensembl/Refseq). Transcript coverage (including UTRs) and coding sequence coverage (excluding UTRs) are supplied for four different depth thresholds. 1X, 5X, 20X and 50X are the default depth thresholds. The user can alter the latter two thresholds under the "Site settings" or under the "Advanced" option in the data upload section.
The exonic coverages can be browsed by clicking on the PLUS (➕) button for each transcript.
The IGV zoom button will zoom on the corresponding region in IGV. You need to add the sample to the IGV to be able to see the corresponding region in the sample (use Open BAM or Open Session)
The gene coverage data can be filtered by using the gene symbol, gene sets, transcript coverage, or coding sequence coverage. Genesets can be added by the user, or you can use the available pre-defined genesets in the SEQ platform.
# FastQC
The FastQC section displays three different quality check data produced by the open-source toolFastQC (opens new window). The following graphs are available:
- The Quality Scores Across All Bases
- The Quality Score Distribution Over All Sequences
- The Sequence Length Distribution