# Somatic Sample Analysis
# Dashboard
Case Information
- (Mandatory) Case-ID/Name: Specific ID/Name created by the user to identify each case
- (Optional) Age (1-150)
- (Optional) Gender (female | male | unspecified)
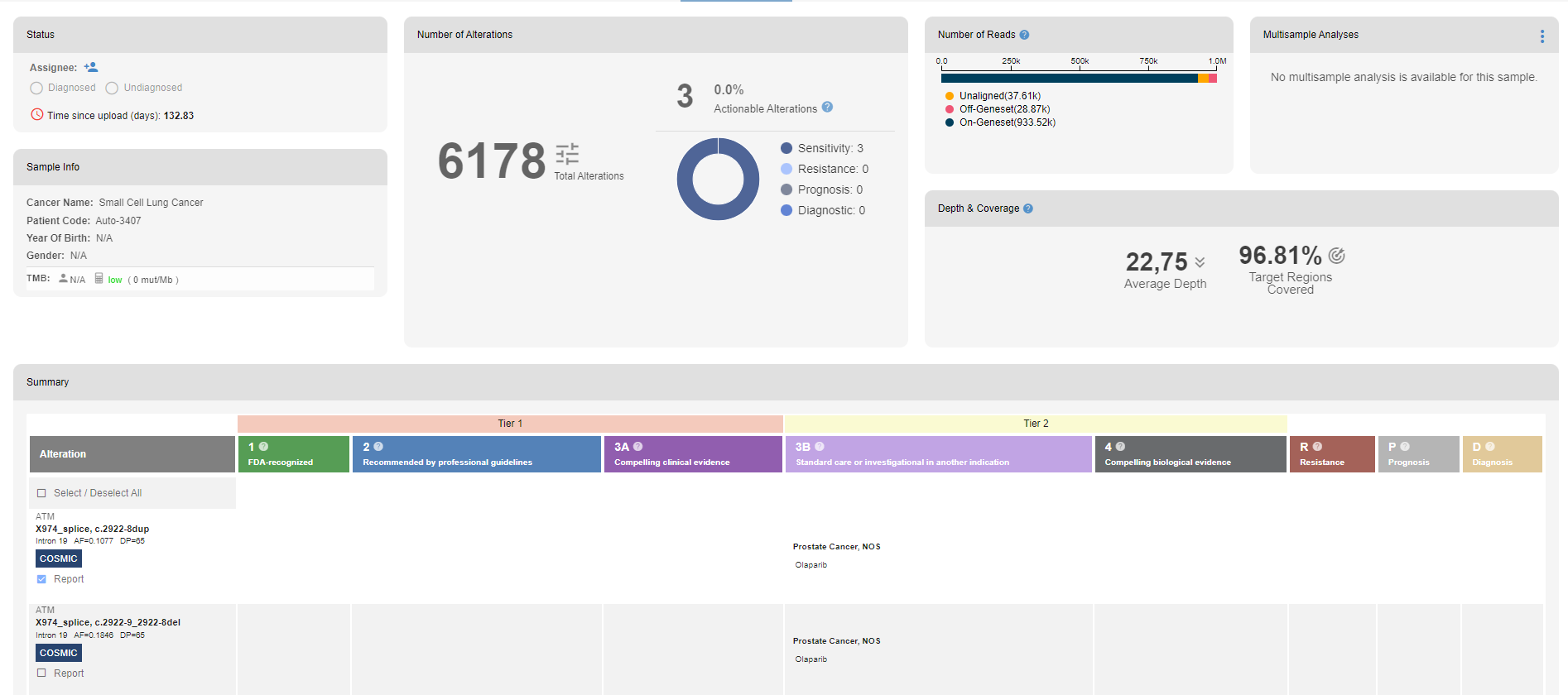
Number of annotations: The number of variant annotations, off-target, and on-target counts are shown here.
Number of reads: The distribution of aligned/unaligned and off/on geneset reads are shown
Het/hom and Ts/tv ratio: The het/hom ratio and the ts/tv ratio of the sample are shown here together with the distribution of the values for all samples in the SEQ platform with the same machine/kit combination.
Target region coverage: The target refers to the regions defined in the kit. The coverage for different sequencing depths is shown for all chromosomes, and per chromosome.
Average depth: The average depth around the kit target regions
% of on target: The target region coverage for all chromosomes with 1X sequencing depth threshold. This is identical to the value illustrated in the target region coverage section.
Summary: In the summary table, drug resistance causing or drug sensitizing alterations and their related drugs are displayed with the level of evidence codes.
- Columns
- Alteration: Name of the gene or biomarker (PD-L1, TMB, MSI, etc.) and how it is altered (e.g. amino acid change, affected transcript position for spliced variant, copy number change, or high TMB)
- Approved for the selected cancer type: List of FDA-approved drugs to treat the diagnosed condition by targeting the alteration or a downstream target. You can see the detailed summary for the drug by clicking each drug name.
- Investigational for the selected cancer type: List of potential drugs that may target specified alterations or a downstream target and are being investigated as a potential therapy for the diagnosed condition. You can see the detailed summary for the drug and links to view the related clinical trials on ClinicalTrials.gov by clicking each drug name.
- Approved for Other Indications: List of drugs that have not been approved to treat the diagnosed condition but have been approved to treat other cancer types.
- Resistance: List of drugs to which at least one of the specified alterations is resistant.
- Columns
# TMB Calculation
Tumor Mutational Burden is defined as non-synonymous SNP mutations per megabase. If the covered region length is less than 1.1 megabases, TMB cannot be calculated and will be seen as 'N/A'. For tumor only samples, TMB is calculated by the total number of non-synonymous and unique/novel SNP variants found in the sample, divided by the covered region on CDS. For tumor/matched-normalsamples, the variants detected in the normal sample are used to eliminate germline mutations.
Please see the table below for detailed parameters used for TMB calculation.
| Parameters | Tumor Only | T/N match |
|---|---|---|
| Minimum Allowed Covered Length | 1.1mb | 1.1mb |
| Minimum Depth | 25 | 50 |
| Minimum Allele Fraction | 0.05 | 0.05 |
| Population Database Allele Filter | 0.1% | - |
| dbSNP filter | Yes | - |
| Chromosome Filter | - | MT |
| Filter Non-coding Regions | Yes | Yes |
| Filter MNVs | - | Yes |
| Filter No PASS Variants | Yes | Yes |
| Use SNV and Indels | Yes | Yes |
| Eligible Region Coverage | 5X | 50X |
# MSI Calculation
MSI calculation is only available for tumor/normal matched analyses. MSIsensor1 (opens new window) is used for MSI calculation.