# Somatic Data Upload
With the SEQ Platform, you can directly upload your files or use the cloud browser to browse and select the files pre-uploaded to your account.
# Direct Data Upload
To upload data for Somatic Analysis, click the PLUS (➕) button on the left menu to open the upload page (disabled in the demo accounts). Select the "Somatic" option.
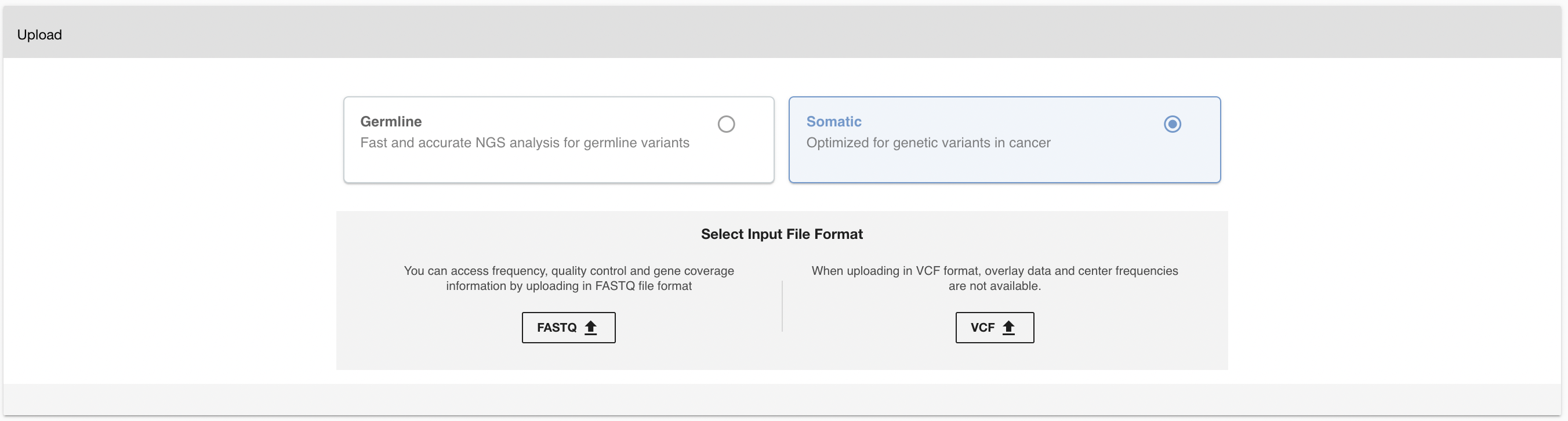
Frequency, quality control, and gene coverage information are available by uploading in FASTQ file format. Overlay data and center frequencies are not available when uploading in VCF format.
# Run Selection
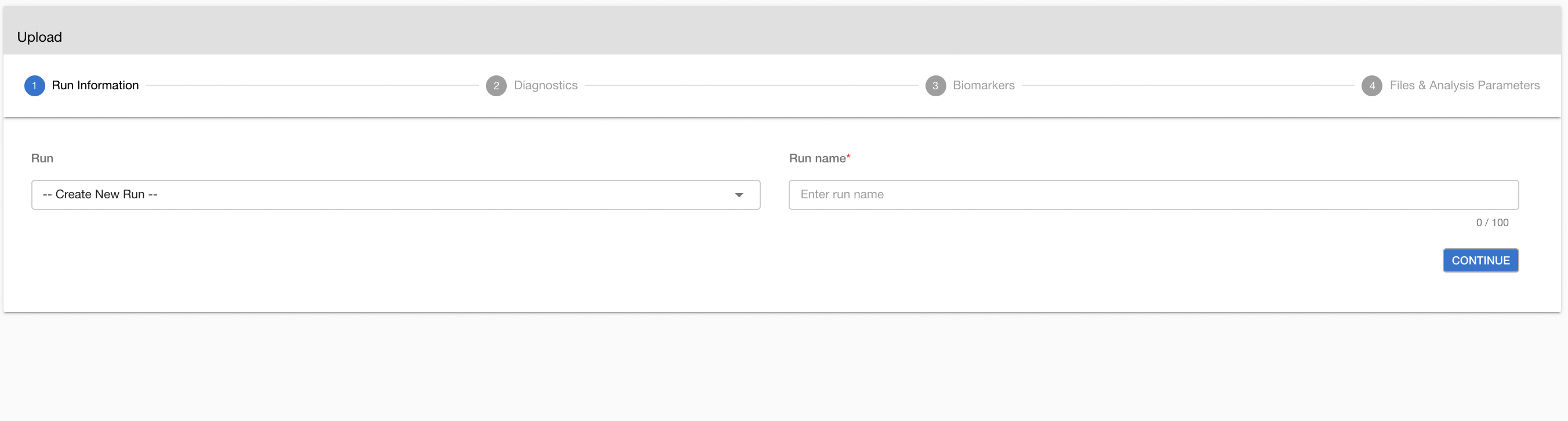
You can upload your samples via two different routes. You can add the samples by creating a new run by selecting “Create New Run” under “Run Name.” Then, you can write the run name in the “Name For New Run” box. You can also upload your samples by adding them to an existing run by selecting the run name where you want to upload the new samples. In this case, you cannot change the name of the run.
Run information is very critical for Copy Number Variation analyses. Therefore, please make sure you organize the samples under the runs in the same way as you process your sample materials. Ideally, a run is a set of samples coming from the same wet-lab and run process (flow cell, etc.).
# Uploading
# Choose the Technology Type
Choose the next-generation sequencing machine associated with the samples. Mixing different technologies in one run is not permitted.
# Choose Your Kit
The SEQ platform has hundreds of different kits predefined in the system. A new kit can be defined with the set of target coordinates and the list of targeted genes. The addition of a new kit typically takes one business day. For kit requests, please contact us at support@genomize.com.
Every kit is associated with a standardized analysis version in SEQ. Probe-based kits, primer-based kits, Illumina & MGI technology, ION torrent technology, germline analysis, or somatic analyses all have a preset analysis version. One sample may be analyzed with multiple analysis versions after upload, although it is not routinely recommended as it may introduce noise to your genotype/phenotype database.
# Choose the Analysis Version
If you are using a probe-capture based library preparation kit, you can use the tumor only or tumor/normalpipeline. Click on the Analysis Version menu and select the pipeline for your analysis.
# Select the files to upload
Click the "Browse" button under the “File” to upload all the files you want to analyze. Make sure that you upload both read files for paired-end reads. See the table below for the supported input file types:
| File Types | Batch Sample Upload | |
|---|---|---|
| Illumina | .fastq.gz, .fq.gz | Not Supported |
| Ion Torrent | .bam | Not Supported |
| MGI | .fastq.gz, .fq.gz | Not Supported |
If you chose Tumor/Normalanalysis pipeline, Normal Files selection will be active. Using these fields, you can select the matched normal file for your tumor samples. Filename formatting is explained below. Formatting rules apply to normal and tumor samples separately. Matching between the tumor and normal samples is not checked. System will assume correct matching tumor and normal files are selected by the user.
# Filename Formatting for File Matching
# Illumina
SEQ Platform can automatically match multiple files from Illumina for a single sample if filenames follow the “Illumina naming convention” e.g.
NA10831_ATCACG_L002_R1_001.fastq.gz
NA10831_ATCACG_L002_R2_001.fastq.gz
The filenames from the Illumina platform are handled as below:
<name_field1>_<name_field2>_<lane_#>_<read_#>_<always_001>.fastq.gz
Name field 1, name field 2, lane #, and read # are used to match the corresponding files correctly. You can alter the name field 1 and the name field 2 without using space and underscore (_) characters. Following this naming convention, you can upload multiple samples (each with several fastq.gz files).
Please check the number of samples and matched files on the confirmation screen.
# MGI
SEQ Platform can automatically match multiple files from MGI for a single sample if filenames follow the "MGI" naming convention” e.g.
V12345678_L01_16_1.fastq.gz
V12345678_L01_16_2.fastq.gz
The filenames from the MGI platform are handled as below:
<flowcell_id>_<lane_#>_<barcode>_<read_#>.fq.gz
Flowcell ID, lane #, barcode, and read # are used to match the corresponding files correctly. You can alter the flowcell ID field without using space
and underscore (_) characters. If you have used more than one barcode for the same sample, you need to rename the file as follows:
Original file names:
sammple1234_L01_16_1.fastq.gz
sammple1234_L01_16_2.fastq.gz
sammple1234_L01_17_1.fastq.gz
sammple1234_L01_17_2.fastq.gz
Altered file names:
sammple1234_L01_16_1.fastq.gz
sammple1234_L01_16_2.fastq.gz
sammple1234_L02_16_1.fastq.gz
sammple1234_L02_16_2.fastq.gz
In this example, the barcode numbers of the last two files are changed to 16, and their lane numbers are increased by 1.
Please check the number of samples and matched files on the confirmation screen.
# IonTorrent
IonTorrent files needs to be unaligned BAM files with .bam extension.
# Diagnostics
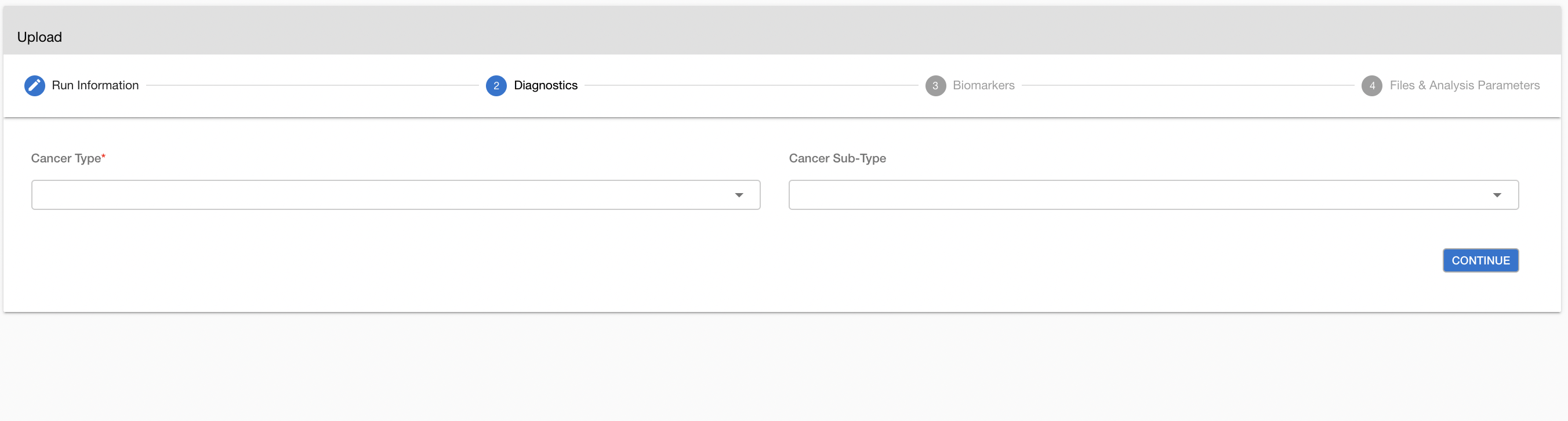
# Cancer Type
Next, you can add the diagnostic information including cancer type and the biomarker status.
- (Mandatory) Cancer Type (e.g. Breast Carcinoma)
- (Optional) Cancer Subtype (e.g. Breast Adenocarcinoma)
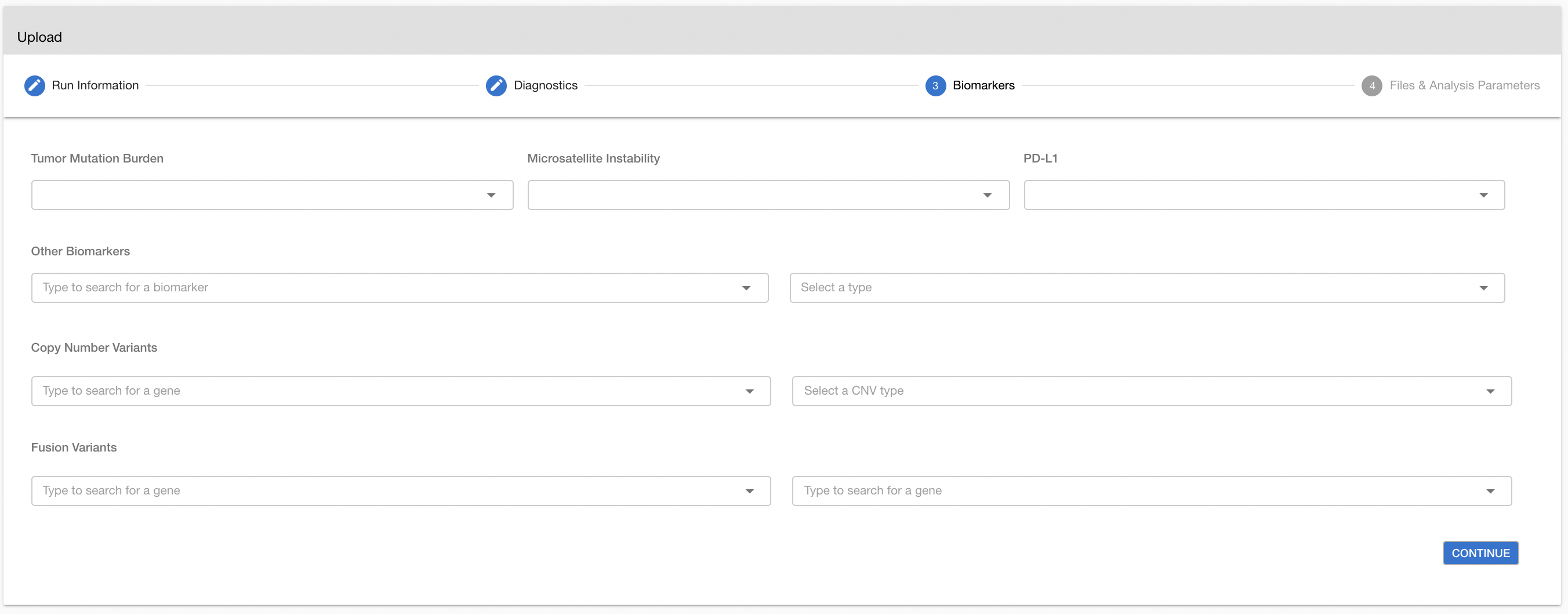
# Biomarkers
(Optional) TMB status: tumor mutation burden status (high | medium | low).The specified TMB status will be used in associating relevant clinical trials and drugs.
(Optional) PD-L1 status: PDL-1 expression status (positive | negative). The PD-L1 status will be used in associating relevant clinical trials and drugs.
(Optional) MSI status: Microsatellite instability status ( high | stable ). The specified MSI status is displayed in the report and used in associating relevant clinical trials and drugs.
(Optional) Other Biormarkers: (positive | negative) Status of clinically relevant biomarkers (e.g. HER2)
(Optional) Copy Number Variations: (amplification | deletion). If there are known CNVs, you can add them by using this parameter to show after analysis.
(Optional) Fusion Variants: If there are known fusion variants, you can add them by using this parameter to show after analysis.
# Small Variant Detection
The SEQ platform has standard analysis versions pre-setup for every kit defined in the system. Calling variants are performed differently in different analysis versions.
# Analysis versions for Tumor/Normal Matched Samples
| Name | Explanation | Alignment/ variant calling | BAM processing | Primer Trimming | Available Genome Versions | Available Platforms |
|---|---|---|---|---|---|---|
| Sentieon BWA-TNhaplotyper2- somatic | Optimized for Exome Samples. | Sentieon BWA / TNhaplotyper2 | MarkDuplicate | N/A | hg38,hg19 | Illumina, MGI |
| Sentieon BWA-TNScope- somatic | Optimized for capture-based somatic kits. | Sentieon BWA / TNScope | MarkDuplicate | N/A | hg38,hg19 | Illumina, MGI |
# Analysis versions for Tumor Only Samples
| Name | Explanation | Alignment/ variant calling | BAM processing | Primer Trimming | Available Genome Versions | Available Platforms |
|---|---|---|---|---|---|---|
| BWA-Freebayes-PCR Dedup-Indel Realignment- somatic | Optimized for capture-based somatic kits. | BWA / Freebayes | PCR Dedup + Indel Realignment | N/A | hg38,hg19 | Illumina, MGI |
| BWA-Freebayes-BamKeser-Indel Realignment-Long Indel Finder- somatic | Optimized for amplicon-based somatic kits. Performs an additional step for long indel alterations. | BWA / Freebayes | Indel Realignment | BamKeser | hg38,hg19 | Illumina, MGI |
| BWA-Freebayes-BamKeser-Indel Realignment- somatic | Optimized for amplicon based somatic kits. | BWA / Freebayes | Indel Realignment | BamKeser | hg38,hg19 | Illumina, MGI |
| Sentieon (ctDNA) BWA-TNscope- somatic | Optimized for ctDNA somatic kits.** | Sentieon BWA / TNScope | MarkDuplicate | N/A | hg38,hg19 | Illumina, MGI |
*BamKeser is our in-house designed and precisely working primer trimming tool.
**For ctDNA somatic kits, the UMI barcode sequence is also required. For assistance, please contact support.
After variant calling, the Genomize SEQ platform processes the resulting VCF file to form a Genomize standard VCF file, which can be downloaded through the platform. The Genomize standard VCF line will have gstd=1 in the info field. Standardization of the VCF file includes the following important steps:
- Minimal variant representation: Some callers produce redundant bases at the left-hand or right-hand side of either alternative or reference allele. This redundancy has to be removed to obtain the correct annotation of variants in the subsequent steps.
- Multi nucleotide polymorphism decomposition: MNPs are decomposed into all possible smaller versions of that variation. This is critical not to miss the annotation of a possible variant in the sample.