# Genomize's AI-Assisted Variant Prioritization
The SEQ Platform offers an AI-assisted variant prioritization (VP) feature to its users.
# Categories
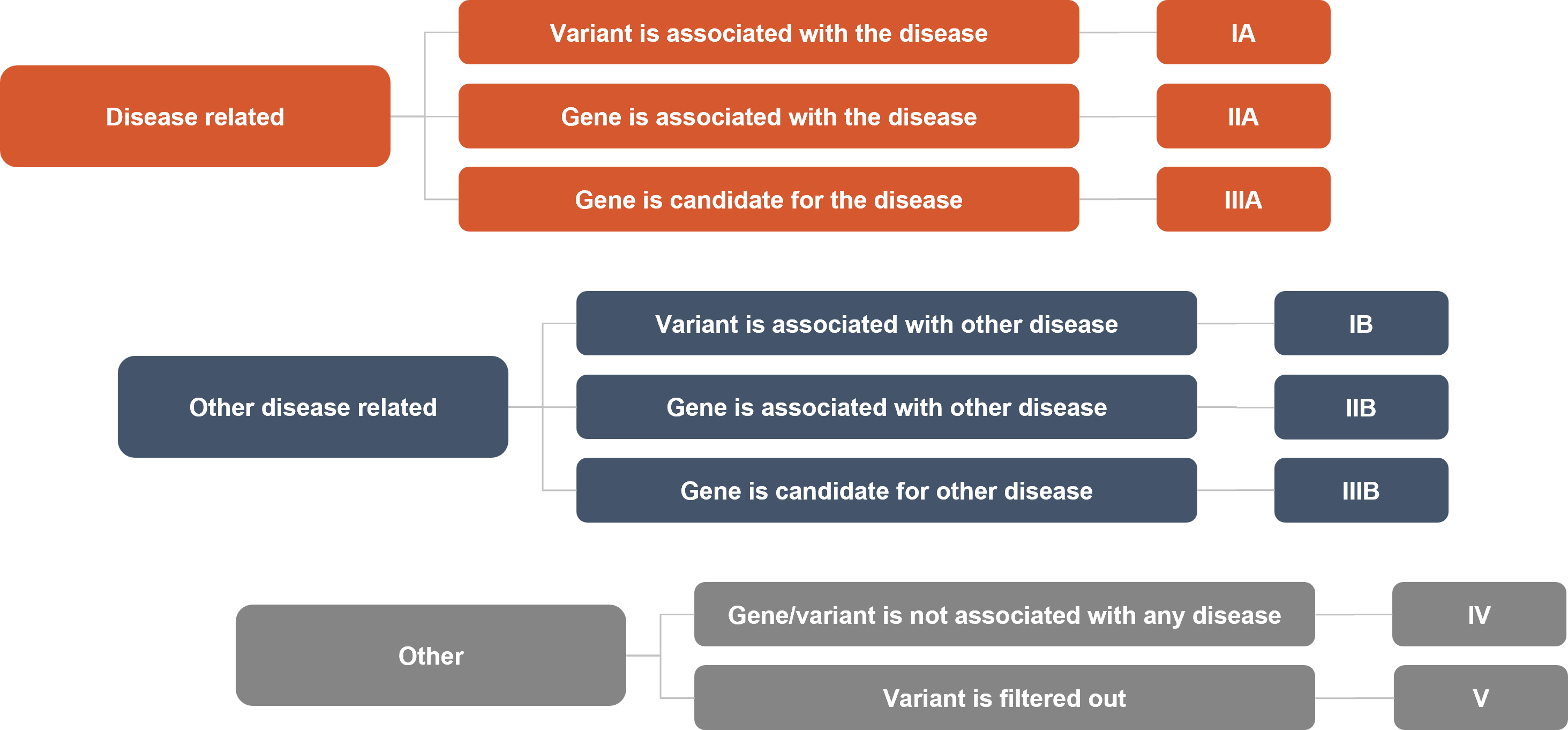
Our proprietary VP algorithm uses the clinical information about the case provided by the user and various information from more than 120 databases and sources to classify each variant observed in the patient into three main and eight sub-categories:
| VP Category | Explanation |
|---|---|
| IA | The variant is known to be associated with the disease input by the user |
| IIA | The gene is associated but the variant is not yet directly associated with the disease |
| IIIA | The variant or the gene is not yet directly associated with the disease but at least one of them is a strong candidate |
| IB | The variant is known to be associated with a disease other than the user input |
| IIB | The gene is associated with a disease other than the user input but the variant is not yet directly associated |
| IIIB | The gene or the variant is a strong candidate for a disease other than the user input |
| IV | The variant may be relevant based on pathogenicity predictions and parameters set by the user (consequence, fraction thresholds, etc.) |
| V | Non-relevant variants or variants with high public or SEQ-community frequencies |
"A" categories are only available when the user provides clinical information during the sample upload. After you have completed the first stage of the upload process (see "Data Upload" section for more details), you will see the "Case Information" screen.
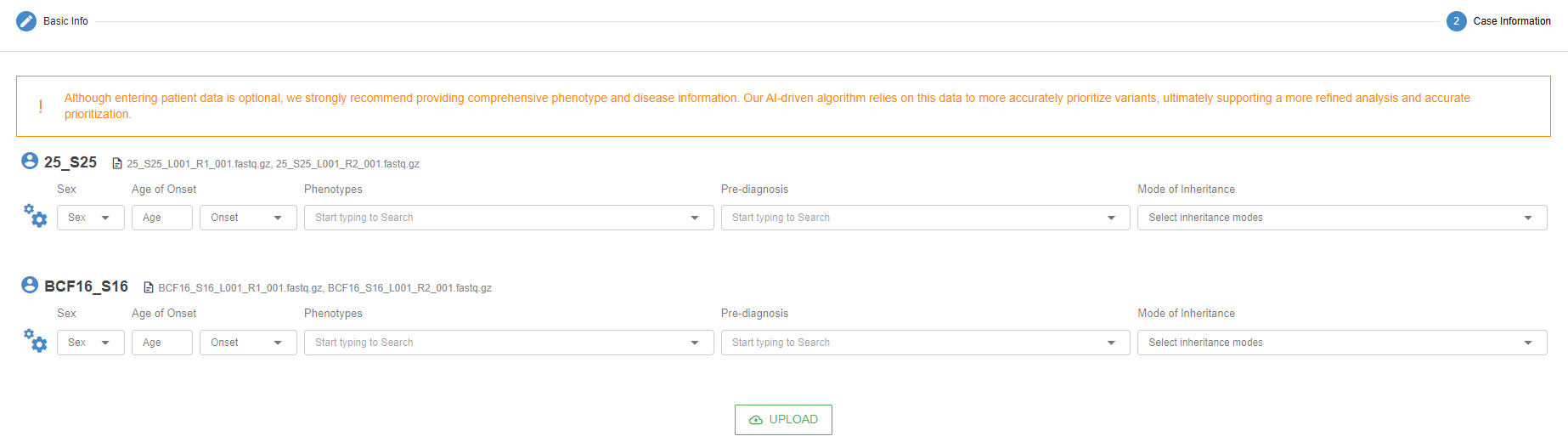
Data entry on this page is optional. Providing the related information, however, will allow the VP AI to categorize variants that could explain the case in the "A" category and allow for shorter and more reliable variant analysis.
By clicking on the "cogs" icon, you can change the default parameters used by the VP for the case.
