# Structural Variant Annotation
# ACMG Classification
Due to currently available guidelines, ACMG classification is only available for DEL and DUP variants according to Riggs et al., 2020 (opens new window).
# Structural Variants Page
When the analysis of your WGS sample is completed, you can click on the "Structural Variants" tab to view and filter detected structural variants in your sample.
Similar to small variants tab, detected structural variants are listed at the top, and the details are shown at the bottom of the page when you click on a variant.
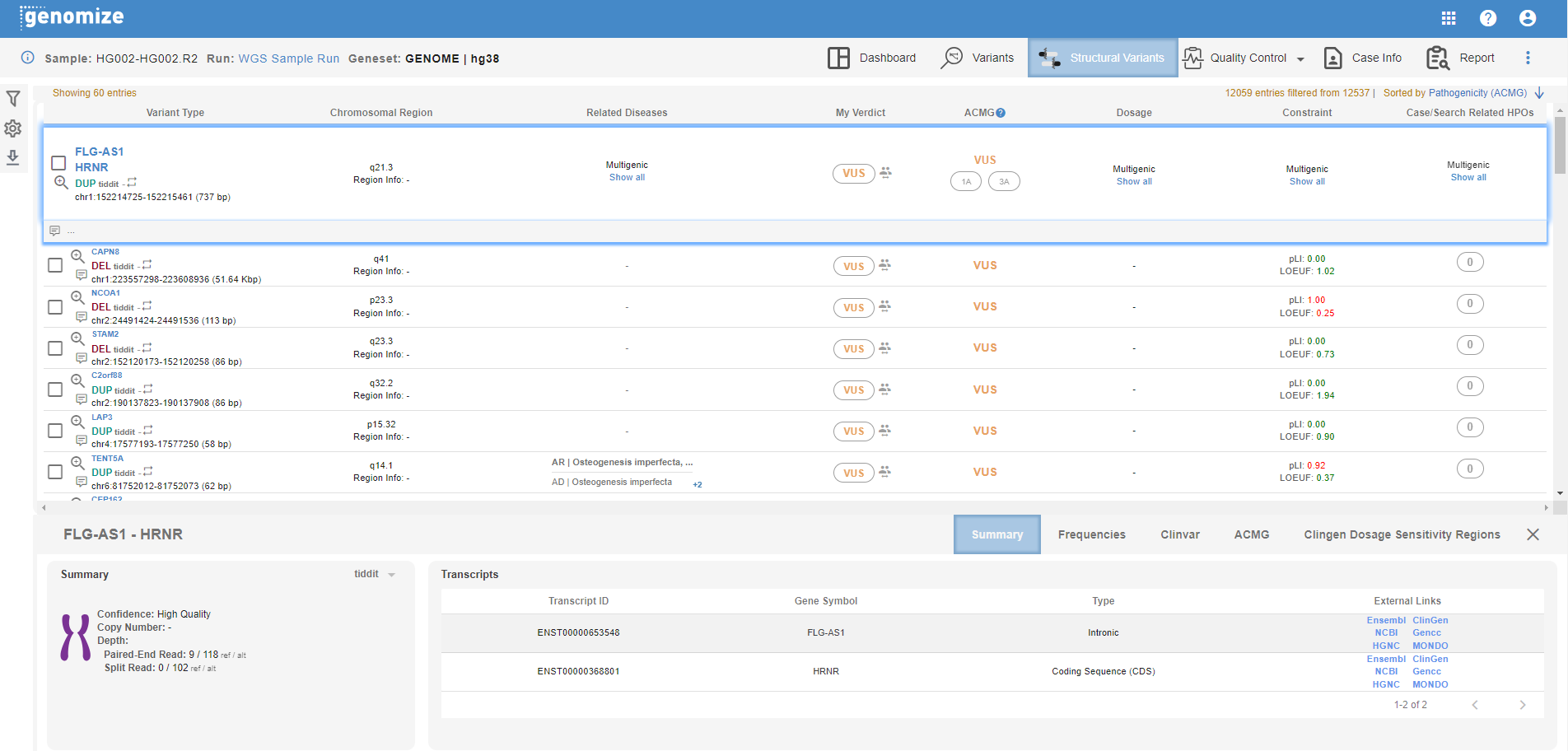
The columns in the list are:
- Variant Type: This field shows the effected gene(s), SNV type, the variant caller that detected the variant, and the chromosomal coordinates and the size of the variant. You can use the report checkbox to include the variant in your report, and use the IGV button to visualize the variant in desktop or web browser IGV depending on your IGV setting.
- Chrosomal Region: The corresponding cytoband is shown here.
- Related Diseases: Disease(s) associated with the affected genes in various databases are shown here.
- My Verdict: You can change the pathogenecity of the variant in this field. The default value is the ACMG pathogenecity calculated by the SEQ Platform. You can also see the statistics for pathogenecity change(s) by other centers by mousing over the people icon.
- ACMG: This field shows the ACMG pathogenecity calculated by the SEQ Platform using the relevant guideline(s). Currently, this information is only available for DEL and DUP SV types.
- Dosage: Haplo insufficiency (HI) and triplosensitivity (TS) information from ClinGen are shown here. For multigenic SVs, click on "show all" to see the list of genes and their corresponding dosage information.
- Constraint: pLI and LOEUF values from GnomAD are shown here. For multigenic SVs, click on "show all" to see the list of genes and their corresponding constraint information.
- Case/Search Related HPOs: Number of matching HPOs is shown here. Click on the variant line to expand the information and see the list of matching HPOs.
At the bottom of your screen you will see the details section. Available tabs and information change based on the SV type:
- Summary: Available for DEL, DUP, STR, and BND. Various information such as zygosity, read counts, supporting read information, etc. are shown here. Available information may change based on SV type.
- Frequencies: Available for DEL, DUP, BND, INS, CPX, and INV. Frequencies in various databases are shown here, if available.
- ClinVar: Available for DEL, DUP, INS, CPX, and INV. List of ClinVar entries and corresponding links are shown here.
- ACMG: Available for DEL and DUP. ACMG pathogenicity calculation, applied evidence codes, and aditional information is shown here.
- ClinGen Dosage Sensitivity Regions: Available for DEL, DUP, BND, INS, CPX, and INV. Additional curation information from ClinGen for the affected genomic regions is shown here, if available.
- Repeat Number Distribution: Available for STR only. The repeat number distribution of the repeat unit is shown here with an interactive graph. You can visualize the distribution for the global data or for available ethnicities.